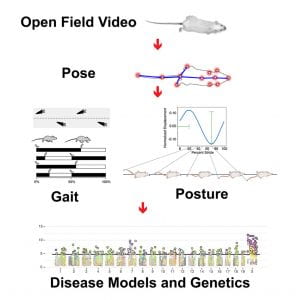
Complete list of publications available from Pubmed and Google Scholar
Vivek Kumar Google Scholar Link Google Scholar
Vivek Kumar My NCBI My NCBI
Selected Publications
Kumar V, Kim K, Joseph C, Kourrich S, Yoo SH, Huang HC, Vitaterna MH, Pardo- Manuel de Villena F, Churchill G, Bonci A, Takahashi JS.
C57BL/6N mutation in Cytoplasmic FMRP interacting protein 2 (Cyfip2) regulates cocaine response.
Science 342: 1508-1512. 2013 (PDF)
News Coverage
Science News
UT Southwestern
Jackson Labs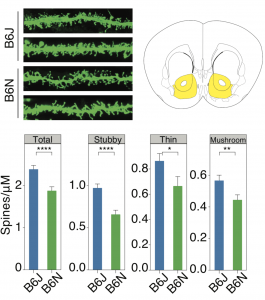
Kumar V, Kim K, Joseph C, Thomas LC, Hong HK, and Takahashi JS.
A Second Generation High Throughput Forward Genetic Screen in Mice to Isolate Subtle Behavioral Mutants.
PNAS 108: Sup 3 15557-15564. 2011 (PDF)
Leinani E. Hession, Gautam S Sabnis, Gary A. Churchill, Vivek Kumar
A machine-vision-based frailty index for mice. Nature Aging. Volume 2(8)-756-766.
https://rdcu.be/cTLJi
News and Views https://rdcu.be/cTLI4
JAX news https://www.jax.org/news-and-insights/2022/September/measuring-frailty-in-mice

Preprints
- Integrated phenotyping platform (Preprint) – https://www.biorxiv.org/content/10.1101/2022.01.13.476229v1
2022
Leinani E. Hession, Gautam S Sabnis, Gary A. Churchill, Vivek Kumar
A machine-vision-based frailty index for mice. Nature Aging. Volume 2(8)-756-766.
https://rdcu.be/cTLJi
News and Views https://rdcu.be/cTLI4
JAX news https://www.jax.org/news-and-insights/2022/September/measuring-frailty-in-mice
A missense mutation in Kcnc3 causes hippocampal learning deficits in mice
Proceedings of the National Academy of Sciences 119 (31), e2204901119
The MABe22 Benchmarks for Representation Learning of Multi-Agent Behavior
2021
Brian Q. Geuther, Asaf Peer, Hao He, Gautam Sabnis, Vivek M Philip, Vivek Kumar
Action detection using a neural network elucidates the genetics of mouse grooming behavior
https://elifesciences.org/articles/63207
Data for paper is here Grooming Data
Brian Geuther, Mandy Chen, Raymond Galante, Owen Han, Jie Lian, Joshy George, Allan I. Pack, Vivek Kumar
High-throughput visual assessment of sleep stages in mice using machine learning
https://academic.oup.com/sleep/article/doi/10.1093/sleep/zsab260/6414386?guestAccessKey=60d3497c-b67a-4953-8130-159130aaed9c
Data for sleep paper is HERE.

Wotton et. al.
Identifying genetic determinants of inflammatory pain in mice using a large-scale gene-targeted screen
https://journals.lww.com/pain/Abstract/9000/Identifying_genetic_determinants_of_inflammatory.97888.aspx
Link to PDF of paper: Identifying_genetic_determinants_of_inflammatory.97888.
2020
Janine M Wotton, Emma Peterson, Laura Anderson, Stephen A Murray, Robert E Braun, Elissa J Chesler, Jacqueline K White, Vivek Kumar
Machine learning-based automated phenotyping of inflammatory nocifensive behavior in mice.
https://doi.org/10.1177/1744806920958596
Data for paper is here Nociception Data
2019
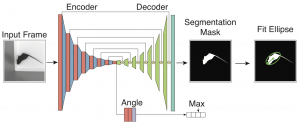
Geuther BQ, Deats SP, Fox KJ, Murray SA, Braun RE, White JK, Chesler EJ, Lutz CM, and Kumar V.
Robust mouse tracking in complex environments using neural networks.
Communications Biology (Nature) 2: 124. 2019.
https://doi.org/10.1038/s42003-019-0362-1
Data for paper is here Tracking Paper Data
Takahashi JS, Kumar V, Nakashe P, Koike N, Huang HC, Green CB, Kim TK.
ChIP-seq and RNA-seq methods to study circaidan control of transcription in mammals.
Methods in Enzymology 551:285-321 2015 (PDF)
Kirkpatrick SL, Goldberg LR, Yazdani N, Babbs RK, Wu J, Reed ER, Jenkins DF, Bolgioni AF, Landaverde KI, Luttik KP, Mitchell KS, Kumar V, Johnson WE, Mulligan MK, Cottone P, Bryant CD.
Cytoplasmic FMR1-Interacting Protein 2 Is a Major Genetic Factor Underlying Binge Eating.
Biological Psychiatry 81(9):757-769 2017 (PDF)
Funato H, Miyoshi C, Fujiyama T, Kanda T, Sato M, Wang Z, Ma J, Nakane S, Tomita J, Ikkyu A, Kakizaki M, Hotta-Hirashima N, Kanno S, Komiya H, Asano F, Honda T, Kim SJ, Harano K, Muramoto H, Yonezawa T, Mizuno S, Miyazaki S, Connor L, Kumar V, Miura I, Suzuki T, Watanabe A, Abe M, Sugiyama F, Takahashi S, Sakimura K, Hayashi Y, Liu Q, Kume K, Wakana S, Takahashi JS, Yanagisawa M.
Forward-genetics analysis of sleep in randomly mutagenized mice.
Nature 539(7629):378-383 2016
Hossain MS, Asano F, Fujiyama T, Miyoshi C, Sato M, Ikkyu A, Kanno S, Hotta N, Kakizaki M, Honda T, Kim SJ, Komiya H, Miura I, Suzuki T, Kobayashi K, Kaneda H, Kumar V, Takahashi JS, Wakana S, Funato H, Yanagisawa M.
Identification of mutations through dominant screening for obesity using C57BL/6 substrains.
Scientific Reports 6:32453 2016
Plikus MV, Van Spyk EN, Pham K, Geyfman M, Kumar V, Takahashi JS, Andersen B.
The Circadian Clock in Skin: Implications for Adult Stem Cells, Tissue Regeneration, Cancer, Aging, and Immunity.
Journal of Biological Rhythms 30:163-182 2015
Stringari C, Wang H, Geyfman M, Crosignani V, Kumar V, Takahashi JS, Andersen, B, Gratton E.
In Vivo single-cell detection of metabolic oscillations in stem cells.
Cell Reports 10: 1-7 2015 (PDF)
Takahashi JS, Kumar V, Nakashe P, Koike N, Huang HC, Green CB, Kim TK.
ChIP-seq and RNA-seq methods to study circaidan control of transcription in mammals.
Methods in Enzymology 551:285-321 2015 (PDF)
Kumar V, Andersen B, Takahashi JS.
Epidermal stem cells ride the circadian wave.
Genome Biology 14: 140-143. 2013 (PDF)
Shimomura K, Kumar V, Koike N, Kim TK, Chong J, Buhr ED, Whiteley AR, Low SS, Omura C, Fenner D, Owens JR, Richards M, Yoo SH, Hong HK, Vitaterna MH, Bass J, Pletcher MT, Wiltshire T, Hogenesch JB, Lowrey PL, Takahashi JS.
Usf1, a suppressor of the circadian clock mutant, reveals the nature of the DNA-binding of the CLOCK:BMAL1 complex in mice.
E-Life 2:e00426. 2013 (PDF)
Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, Kornblum I, Kumar V, Koike N, Xu M, Nussbaum J, Liu X, Chen Z, Chen ZJ, Green CB, and Takahashi JS.
Competing E3 Ubiquitin Ligases Govern Circadian Periodicity by Degradation of CRY in Nucleus and Cytoplasm.
Cell 152(5): 1091-1105. 2013.
Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS.
Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals.
Science 338(6105): 349-354. 2012.
Geyfman M, Kumar V, Liu Q, Ruiz R, Gordon W, Espitia F, Cam E, Millar SE, Smyth P, Ihler A, Takahashi JS, Andersen B.
Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis.
PNAS 109(29):11758-63, 2012.
Kumar V and Takahashi JS.
PARP around the Clock.
Cell 142(6): 841-843. 2010 (PDF)
Shimomura K, Lowrey PL, Vitaterna MH, Buhr ED, Kumar V, Hanna P, Omura C, Izumo M, Low SS, Barrett, RK, LaRue SI, Green CB, Takahashi JS.
Genetic suppression of the circadian Clock mutation by the melatonin biosynthesis pathway.
PNAS 107(18): 8399-8403. 2010.
Chen R, Schirmer A, Lee Y, Lee H, Kumar V, Yoo SH, Takahashi JS, Lee C.
Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism.
Molecular Cell 36(3):417-30. 2009.
Lin KK, Kumar V, Geyfman M, Chudova D, Ihler AT, Smyth P, Paus R, Takahashi JS, Andersen B.
Circadian clock genes contribute to the regulation of hair follicle cycling.
PLoS Genetics Jul;5(7):e1000573. 2009.
Takahashi J.S., Shimomura K., Kumar V.
Searching for genes underlying behavior: lessons from circadian rhythms.
Science 322(5903):909-12. 2008.
Siepka SM, Yoo, SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS.
Circadian mutant Overtime reveals F-box protein FBXL3 regulation of Cryptochrome and Period gene expression.
Cell 129(5): 1011-1023. 2007.
Kumar V, Carlson JE, Ohgi KA, Edwards TA, Rose DW, Escalante CR, Rosenfeld MG, Aggarwal AK.
Transcriptional corepressor CtBP is an NAD(+)- regulated dehydrogenase.
Molecular Cell 10(4):857-69. 2002 (PDF)
Sugihara TM, Kudryavtseva EL, Kumar V, Horridge JJ, Andersen B.
The POU domain factor Skin-1a represses the keratin 14 promoter independent of DNA binding. A possible role for interactions between Skn-1a and CREB-binding protein/p300.
Journal of Biological Chemistry 276(35): 33036-44. 2001.
Jepsen K, Hermanson O, Onami TM, Gleiberman AS, Lunyak V, McEvilly RJ, Kurokawa R, Kumar V, Liu F, Seto E, Hedrick SM, Mandel G, Glass CK, Rowe DW, Rosenfeld MG.
Combinatorial roles of the nuclear receptor corepressor in transcription and development.
Cell 102(6):653-63. 2000.
Fonstein M, Koshy EG, Kumar V, Mourachov P, Nikolskaya T, Tsifansky M, Zheng S, Haselkorn R.
Rhodobacter capsulatus SB1003.
In Bacterial Genomes: Physical Structure and Analysis, de Bruijn, Lupski, and Weinstock (eds) 1998.
Kumar V, Fonstein M, Haselkorn R.
Bacterium genome sequence.
Nature 381:653-4. 1996 (PDF)
[/et_pb_text][/et_pb_column][/et_pb_row][/et_pb_section]